The surfaces of grapevines host a rich and dynamic microbiome that can influence vine health, grape quality, and ultimately, wine style. These microbial communities vary between wine regions and are shaped by grape variety, climate, geography, and vineyard practices. One well-adapted yeast species is of particular interest as discussed in the study “Genetic and phenotypic diversity of wine-associated Hanseniaspora species” in FEMS Yeast Research and on this #FEMSmicroBlog by Cristobal Onetto. #FascinatingMicrobes
The vineyard and grape microbiome: a complex mixture of fungi and bacteria
By harvesting grapes, the naturally occurring mix of fungi and bacteria on their surfaces is carried into the winery. Throughout the entire wine-making process, these microbial communities continue to play vital roles in creating the final product.
In spontaneous (‘wild’) fermentations, no commercial yeast is added. The native grape microbiota initiates fermentation and thus contributes to the complexity of the final wine. As the grape juice ferments, a succession of microbial interactions unfolds.
While many species are involved, some are clearly better adapted to compete in this environment. A standout example is yeast from the genus Hanseniaspora, particularly the species Hanseniaspora uvarum. These lemon-shaped yeasts often dominate the early stages of fermentation, actively influencing fermentation performance and the wine’s chemical profile.
Eventually, Saccharomyces cerevisiae overtakes them since it better tolerates high ethanol concentrations and anaerobic conditions. Still, the early influence of Hanseniaspora can leave a lasting impact on the style of wine as it produces distinctive metabolites and influences fermentation kinetics. Despite their prevalence, the traits that give Hanseniaspora a competitive advantage remain poorly understood.
The study “Genetic and phenotypic diversity of wine-associated Hanseniaspora species” in FEMS Yeast Research aimed to understand what makes some Hanseniaspora strains more fit than others in the fermentative environment and how much diversity exists within wine-associated Hanseniaspora strains.
Exploring wine-associated Hanseniaspora
To explore these questions, scientists from the Australian Wine Research Institute sequenced the genomes of over 150 Hanseniaspora isolates collected from wineries across Australia. These isolates grouped into two major evolutionary lineages. Remarkably, the genome structure between these lineages differed substantially.
In particular, the lineage containing Hanseniaspora uvarum had lost many genes involved in core cellular processes like RNA splicing. Since these functions are considered essential, the researchers questioned whether the gene loss might have resulted in less regulatory control and faster growth.
They also examined 83 strains of Hanseniaspora uvarum in greater detail and uncovered substantial variation in genes linked to pesticide resistance, nutrient uptake, and stress tolerance. Some of these gene variants appear to be adaptations to vineyard and winery conditions, such as high sugar concentrations, sulfur dioxide, or pesticides. These traits may help the yeast dominate both the surface of grapes and during the early stages of fermentation.
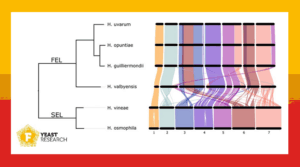
To support these genomic findings, the scientists tested the ability of a large set of strains to grow under common fermentation-related conditions. The results revealed striking differences in stress tolerance, including resistance to ethanol, osmotic stress, and low temperatures. These differences are not only present between species, but also among strains of the same species.
These phenotypic differences mirror the genetic variation observed and reinforce the idea that not all Hanseniaspora strains are alike. This study showed the microbial diversity of the wine microbiome. Beyond winemaking, this work positions Hanseniaspora as a powerful model system for studying how microorganisms evolve and adapt to agricultural fruit environments.
The pronounced loss of genes associated with essential cellular processes in Hanseniaspora suggests novel evolutionary strategies and competitive dynamics, where streamlined genomes may enhance competitiveness. These results may have broader implications for understanding microbial evolution in agricultural and industrial settings.
- Read the article “Genetic and phenotypic diversity of wine-associated Hanseniaspora species” by Onetto et al. in FEMS Yeast Research (2025).

Dr. Cristobal Onetto is a Senior Research Scientist at the Australian Wine Research Institute with over 13 years of experience in the wine industry, spanning roles as both a winemaker and research scientist. His expertise encompasses a broad range of wine science fields, including fermentation performance, microbiology, and the genetics of yeast, bacteria, viticultural pathogens, and grapevines.
About this blog section
The section #FascinatingMicrobes for the #FEMSmicroBlog explains the science behind a paper and highlights the significance and broader context of a recent finding. One of the main goals is to share the fascinating spectrum of microbes across all fields of microbiology.
| Do you want to be a guest contributor? |
| The #FEMSmicroBlog welcomes external bloggers, writers and SciComm enthusiasts. Get in touch if you want to share your idea for a blog entry with us! |
