As 2025 comes to an end, it is a great time to look back at the diverse microbes that made their debut to the scientific community this year. In this first post of the “New Microbes Discovered in 2025” series, Sarah Wettstadt highlights some of the most fascinating bacteria identified in 2025. As always, the list of publications on new bacterial species is too long for one blog. That’s why this #FEMSmicroBlog aims to give a broad overview of new bacteria from environmental sources — featuring fascinating shapes and metabolic functions. #NewMicrobes
A marine glass sponge contains a diverse microbial community
Marine sponges are ancient organisms that emerged 600–890 million years ago. Over time, they developed complex relationships with microbial symbionts, which can constitute up to 40% of the sponge biomass.
The study “Survival strategies for the microbiome in a vent-dwelling glass sponge from the middle Okinawa Trough” investigated the microbial community associated with a glass sponge from a hydrothermal vent. The scientists retrieved 11 previously unknown prokaryotic metagenome-assembled genomes from the Thermoproteota, Nitrospinota, Omnitrophota, and Pseudomonadota genera, with Pseudomonadota being the most dominant bacterial phylum.

While they classified two new genera related to Omnitrophota and Pseudomonadota, newly identified species within the Pseudomonadota genus were highly diverse. One species, from the family Methylomonadaceae, was related to a methane-oxidising sponge symbiont.
Others were related to sulphur-oxidising bacteria found in hydrothermal ecosystems, while two more contained genes for thiosulphate oxidation. Of these, one isolate from the Halieaceae family harboured genes involved in denitrification, and the other carried genes for the dissimilatory nitrate reduction pathway.
Also, genes involved in the core carbon metabolism showed notable variation. Hence, this study highlights the metabolic diversity of the sponge-associated microbiome, with many questions still to be answered.
Rice bacteria increasing their surface area
Bacteria evolved their cell shapes for optimal nutrient acquisition, motility, and stress resistance. For example, increasing the surface-area-to-volume ratio is a well-known strategy to ensure survival under nutrient-limited conditions.
The study “Unusual Morphological Changes of a Novel Wrinkled Bacterium Isolated from the Rice Rhizosphere Under Nutrient Stress” identified a Gram-negative, aerobic bacterium with a novel adaptive strategy to nutrient starvation. The scientists proposed a new genus with the type species Rugositalea oryzae, related to Pseudorhodoplanes sinuspersici, belonging to the order Rhizobiales.
When grown in high-nutrient media, Rugositalea oryzae produced smooth, rod-shaped cells. Grown in low-nutrient media, however, they showed wrinkles and transformed into shapes similar to fusilli pasta. The hypothesis is that by producing these wrinkles, cells can hold more transporters in the cell membrane to take up more nutrients from the environment.

Interestingly, the bacterium contains an increased number of genes related to cell cycle control, cell division, peptidoglycan biosynthesis, and cell wall remodelling, all of which influence cell shape and growth. These could give a first clue to understanding the molecular mechanisms behind this novel cell shape.
The human skin microbiome shows high antibiotic resistance
We still lack a full understanding of how commensal skin microbes interact with the host and other microbes to maintain skin health. Despite Corynebacterium species being a major component of the human skin microbiome, research has focused on other bacterial players.
The study “Revealing the diversity of commensal corynebacteria from a single human skin site” collected axillary swabs from four volunteers and enriched them for corynebacteria on selective agar. The researchers identified 30 genetically distinct isolates spanning seven Corynebacterium species. This included Corynebacterium gottingense, not previously associated with human skin, as well as two novel species—Corynebacterium axilliensis, related to Corynebacterium singulare, and Corynebacterium jamesii, related to Corynebacterium jeikeium.
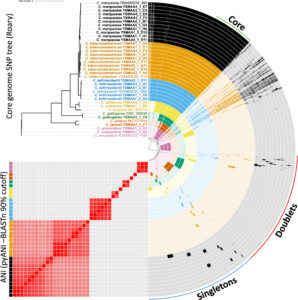
Since bacteria leverage antibiotic resistance to gain competitive advantage in their niche, the study focused on such genes. Interestingly, one Corynebacterium axilliensis isolate contained genes conferring resistance to chloramphenicol, streptomycin, and kanamycin. A second isolate of this species lacked these genes, indicating recent acquisition.
In comparison, the Corynebacterium jamesii isolate was resistant to penicillin G despite lacking a β-lactamase. This species likely uses another mechanism to resist penicillin G. Overall, this study shed light on the commensal players of the skin microbiome and their abilities to survive in this environment.
A new bacterium from space
In April 2018, scientists isolated four Gram-stain-positive bacterial strains from the inside walls of the International Space Station. All four strains were related to Microbacterium paraoxydans, belonging to the family Microbacteriaceae under the order Micrococcales.
While not discovered this year, the 2025 study “Genomic characterisation of Microbacterium meiriae sp. nov., a novel bacterium isolated from the International Space Station” described these strains and their phenotypic and phylogenetic features. Genomic and phylogenomic analysis revealed that these isolates represent distinct strains of the new species Microbacterium meiriae.
Functional genome analysis showed that these strictly aerobic, mesophilic rods contain unique proteins associated with transcription, defense—including toxin-antitoxin systems—and metabolism. Microbacterium meiriae also carries a gene conferring tetracycline resistance that is 82% sequence identical to the gene of Microbacterium paraoxydans.
Given its ecological resilience, this new species may be clinically relevant. This study highlights the importance of understanding microbial adaptation and its implications for prolonged human missions in space.
Welcome to the new bacteria identified in 2025
Here, we discussed a few examples of the remarkable bacterial diversity discovered in 2025 — from deep hydrothermal vents to the soil and our own skin, and even outer space. Each new species reveals unique adaptation mechanisms and metabolic capabilities that expand our understanding of microbial life.
Stay tuned for the next posts in this series, where we’ll explore newly identified archaea, fungi, and viruses that made their debut this year.

Dr Sarah Wettstadt is a microbiologist-turned science writer and communicator writing for professional associations, life science organisations and researchers from the biological sciences. She runs the blog BacterialWorld to share the diverse and colourful activities of microbes and bacteria, based on which she co-published the colouring book “Coloured Bacteria from A to Z“. As science communication manager for the Scientific Panel on Responsible Plant Nutrition and blog post commissioner for the FEMSmicroBlog, Sarah writes about microbiology and environmental topics for various audiences. To help scientists improve their science communication skills, she co-founded SciComm Society. Prior to her science communication career, Sarah completed a PhD at Imperial College London, UK, and a postdoc at the CSIC in Granada, Spain. In her non-scicomm time, she enjoys playing beach volleyball on the sunny beaches in Spain or travelling the world.
About this blog section
Each year, the #NewMicrobes series for the #FEMSmicroBlog explores new species discovered throughout the year. Through several posts, we highlight the microbial diversity across all kingdoms by showcasing newly identified bacteria, viruses, fungi, and archaea.
| Do you want to be a guest contributor? |
| The #FEMSmicroBlog welcomes external bloggers, writers and SciComm enthusiasts. Get in touch if you want to share your idea for a blog entry with us! |

