Microbiology Researchers Test Five AI Tools
The AI boom is transforming every part of how we work, research included. With so many tools now marketed to scientists, we asked two microbiologists, Antonella Colque and Nurdana Orynbek, to test a selection of AI tools built for scientific publishing and research. Antonella and Nurdana are both active researchers in the field and FEMS science communication experts, making them the ideal testers for this review.
The goal? To see how well these tools perform in real research tasks, from searching papers and comparing experiments to identifying research gaps.
Here’s what worked for them, what didn’t, and how they suggest you use it in your own research.
Jump straight to the tools
Quick Comparison Table
| Stage of research | Best tool | Why it works? |
| Exploring a new topic | Perplexity | Combines live web + academic data to return concise summaries with inline citations |
| Comparing studies / systematic review | Elicit | Extracts structured data from papers and builds ready-to-use comparison tables |
| Identifying research gaps / hypothesis building | FutureHouse | Uses domain-specific agents to suggest patterns and research opportunities |
| Summarizing and writing | SciSpace | Integrates search, “chat with PDF,” report writing, and citations in one place |
| General brainstorming / refinement | ChatGPT | Fast, flexible dialogue with detailed explanations and references |
Tool Spotlights
Perplexity
How it works
Perplexity combines search engine with an AI assistant. It provides concise answers with clickable citations so you can verify the source. “Research Mode” aggregates and summarizes multiple papers at once, pulling data from Semantic Scholar and other academic databases.
Testing
▶ Nurdana’s Test (Click to expand)
The prompt:
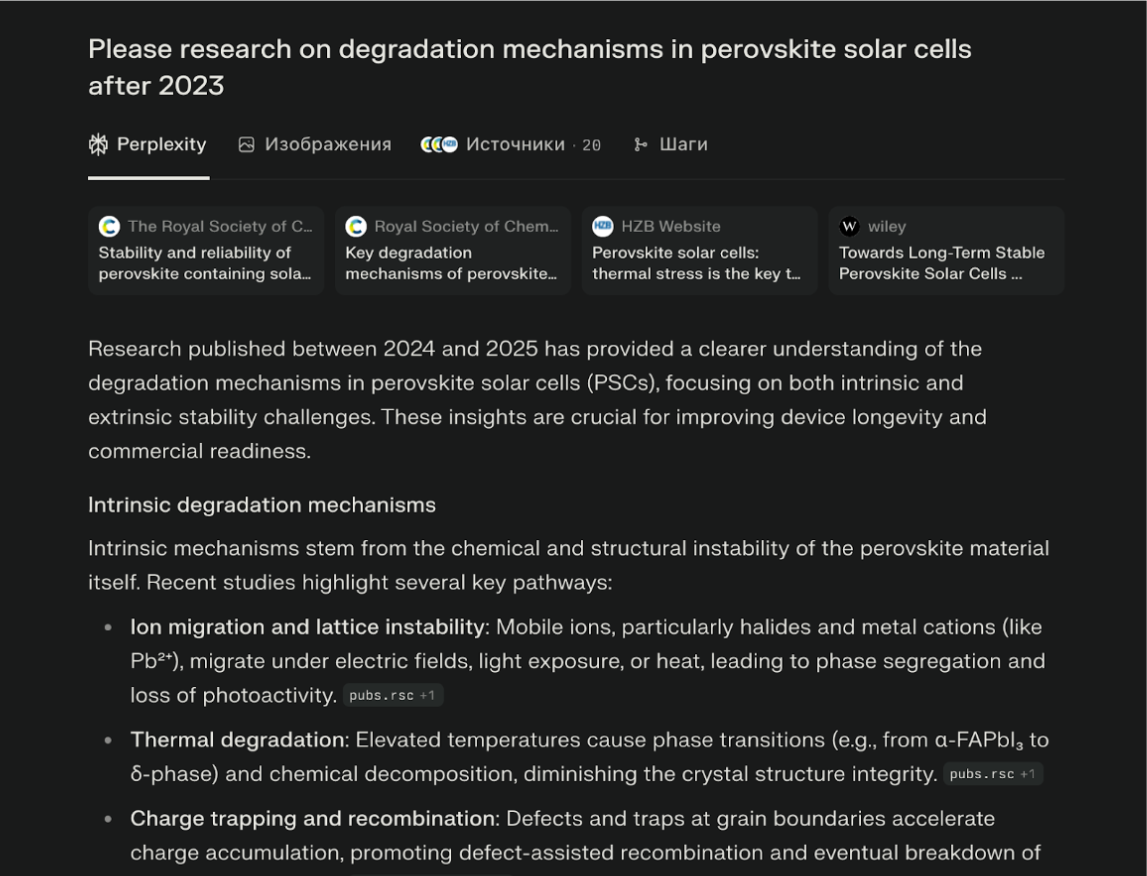
Please research on the degradation mechanisms in perovskite solar cells after 2023
The response:
It summarized the key issues (moisture, ion migration) and linked directly to Nature Energy and Advanced Materials papers.

▶ Antonella’s Test (Click to expand)
The prompt:
Compare different transformation methods in bacteria.
The response:
During the first attempt, Perplexity buffered and unfortunately did not return a result.
When trying again at a later date, Perplexity successfully returned answers, suggesting the earlier failure was likely a temporary system issue. While she was more satisfied with the output this time around, she noted a strange UI behavior: some answers included “links to images.png,” but clicking them only opened a reference to her own query, making the citations feel unreliable or misleading.
She also confirmed that the free version only allows three “Pro” searches per day, after which it downgrades to basic search until the next day.
Pros & cons
| What worked | What didn’t |
|---|---|
|
|
Microbiologist’s takeaway:
While Perplexity offers many use cases, real-time information, article summarization, research support, creative generation, and tailored guidance, our testers found it most useful for quick literature discovery and concise, source-linked summaries.
Nurdana noted, “It feels closer to an interactive Google Scholar, showing sources inline and giving instant access to full texts when available. For exploratory reviews, it surfaces recent papers effectively and often points to primary literature I might have missed.”
Antonella, however, had a different experience. She encountered technical issues during her first attempt, and on her second try found the results broadly similar to ChatGPT and therefore, nothing particularly impressive.
Elicit
How it works
Made specifically to support academic research, Elicit searches scholarly databases semantically (not just by keywords) then extracts structured information, such as methods, results, and key parameters, into a neat table.
Testing:
▶ Nurdana’s Test (Click to expand)
The prompt:
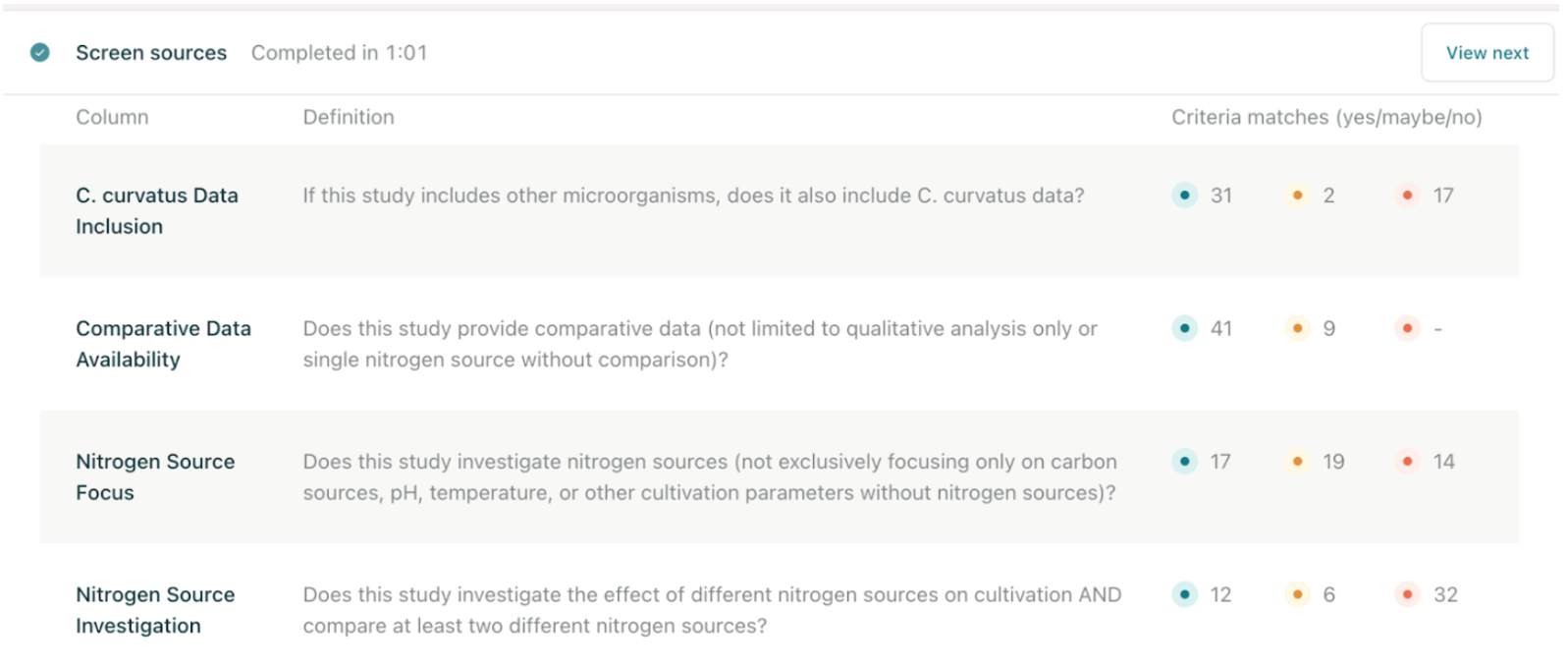
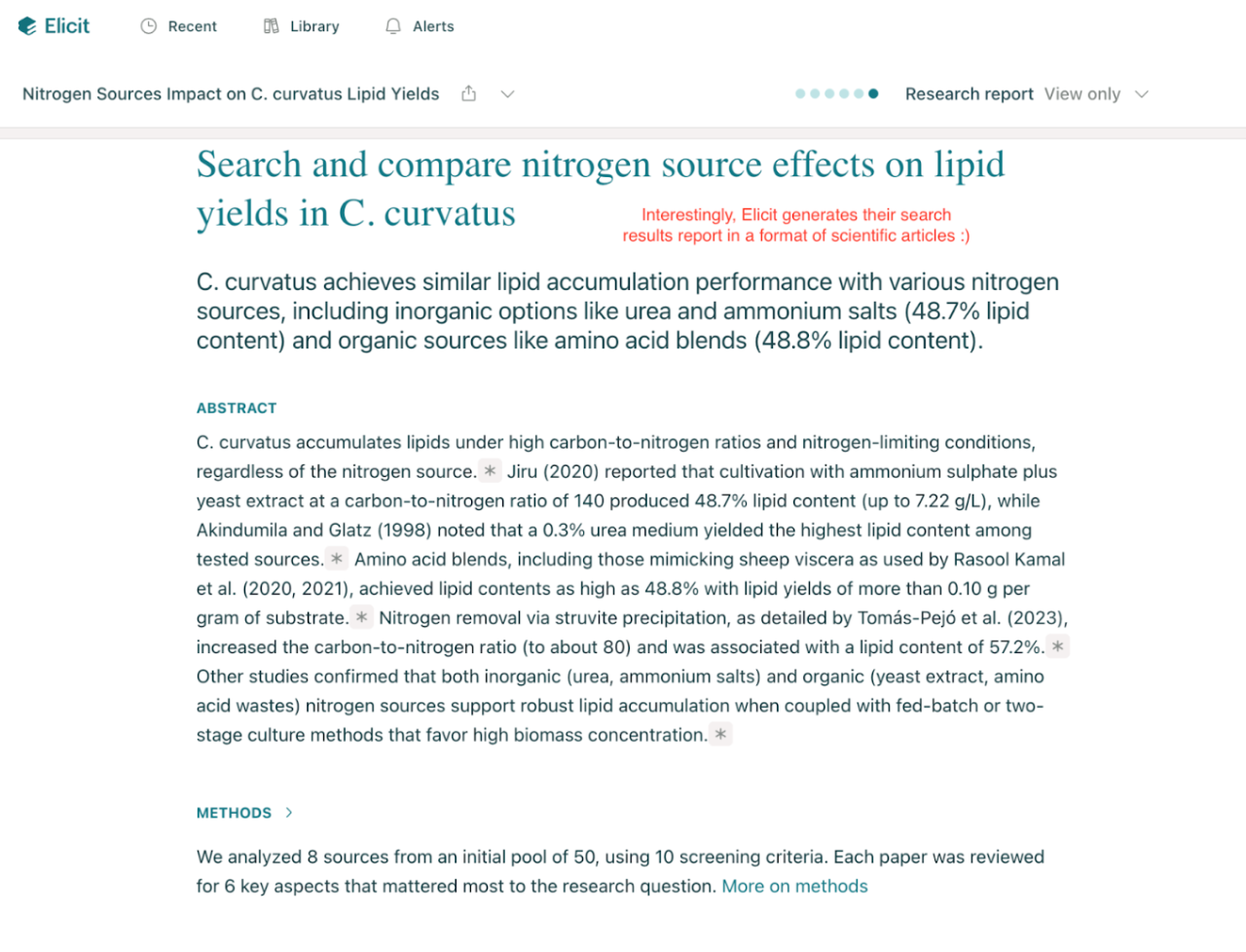
Nitrogen source effects on lipid yields in C. curvatus.
The response:
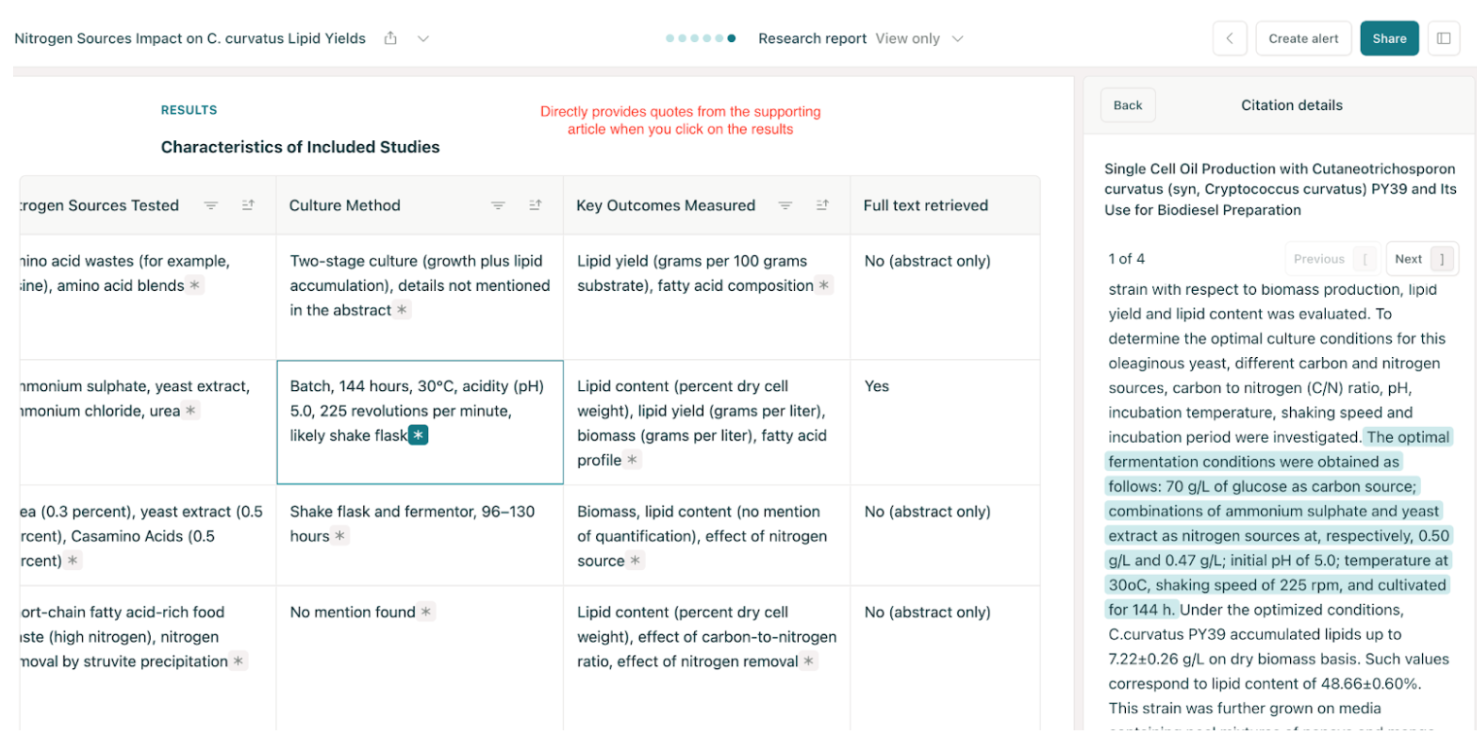
Elicit listed around 8 studies with their nitrogen types, concentrations, and lipid yields. It clearly showed that ammonium sulphate and urea often gave higher yields under nitrogen limitation, something that would’ve taken hours to find manually. I liked how it quotes text directly from the papers, which helps with verification.
▶ Antonella’s Test (Click to expand)
Antonella did not test Elicit.
Pros & cons
| What worked | What didn’t |
|---|---|
|
|
Microbiologist’s Takeaway
Like Perplexity, Elicit appears to be most useful in the early stages of the research process. Nurdana noted that “instead of opening every paper, it collects key data points—such as experimental setup, metrics, and findings—and displays them side by side. This makes it easier to spot patterns or gaps.” She also found it “more reliable and research-oriented than most AI tools, as it prioritizes factual extraction over creative summarization.”
FutureHouse
How it works
FutureHouse is a part of a new wave of AI-for-science projects. It hosts specialized agents (“Crow,” “Falcon,” “Phoenix,” and “Owl”) designed for different kinds of scientific reasoning, from quick searches to chemistry analysis, and hypothesis discovery.
Testing
▶ Antonella’s Test (Click to expand)
The prompt:
Has anyone explored the role of exopolisaccharides like alginate, in airway ALI models?
The response:
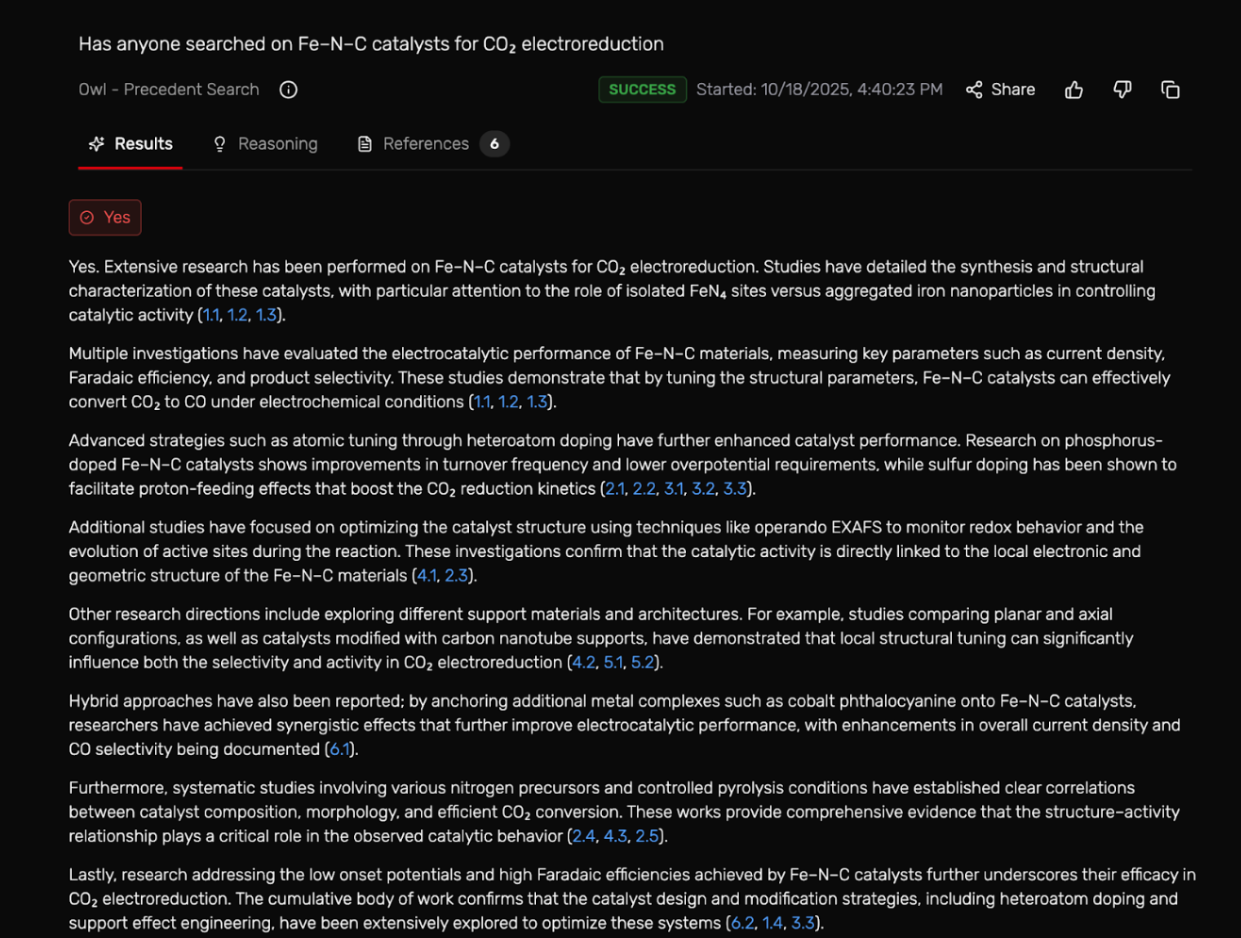
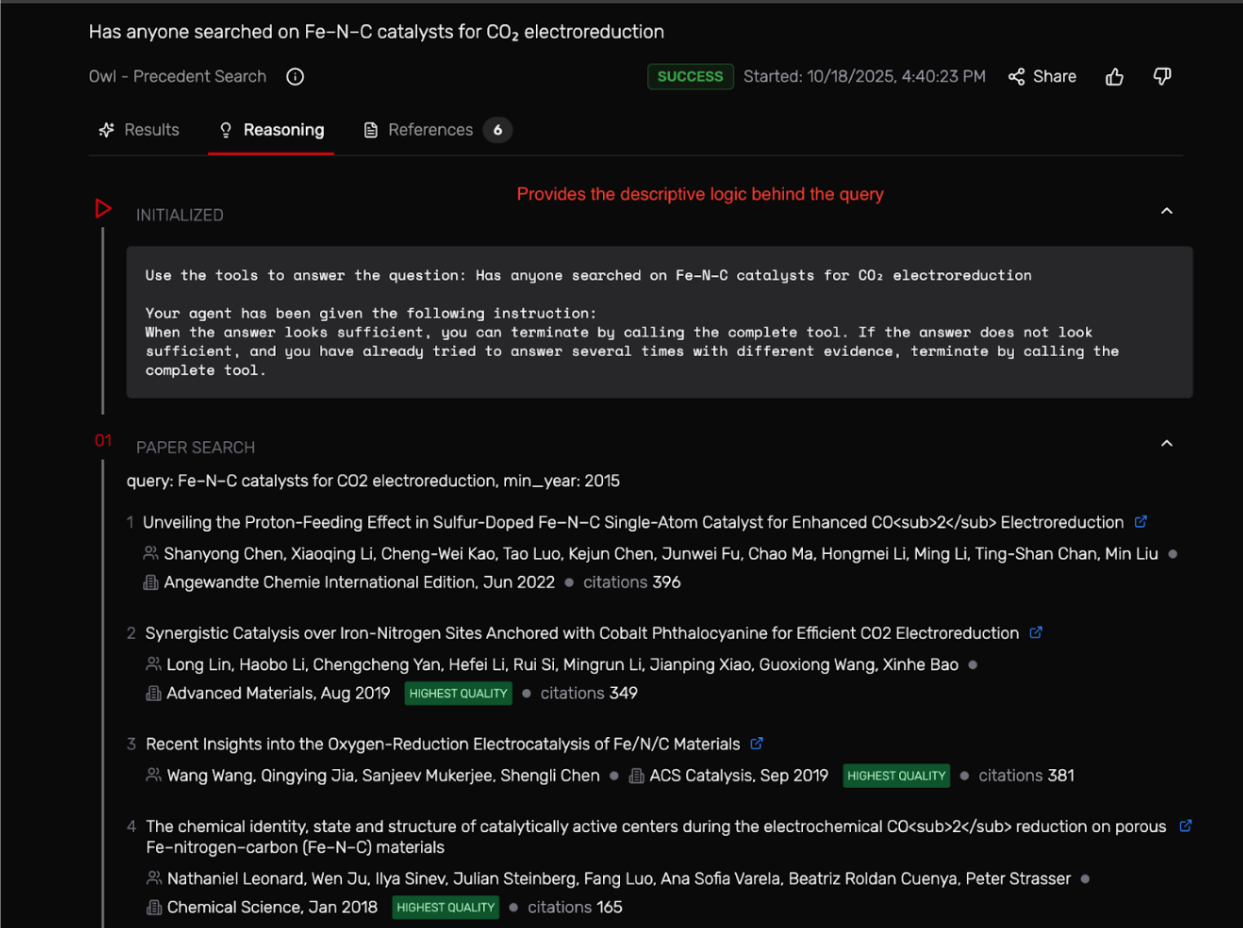
The results window shows the reasoning process behind the search by:
- Displaying relevant publications with clickable links
- Gathering evidence from these papers to identify the most relevant ones
- Repeats the search changing some of the wording and filtering for new papers (min_year 2015)
- It does another query search
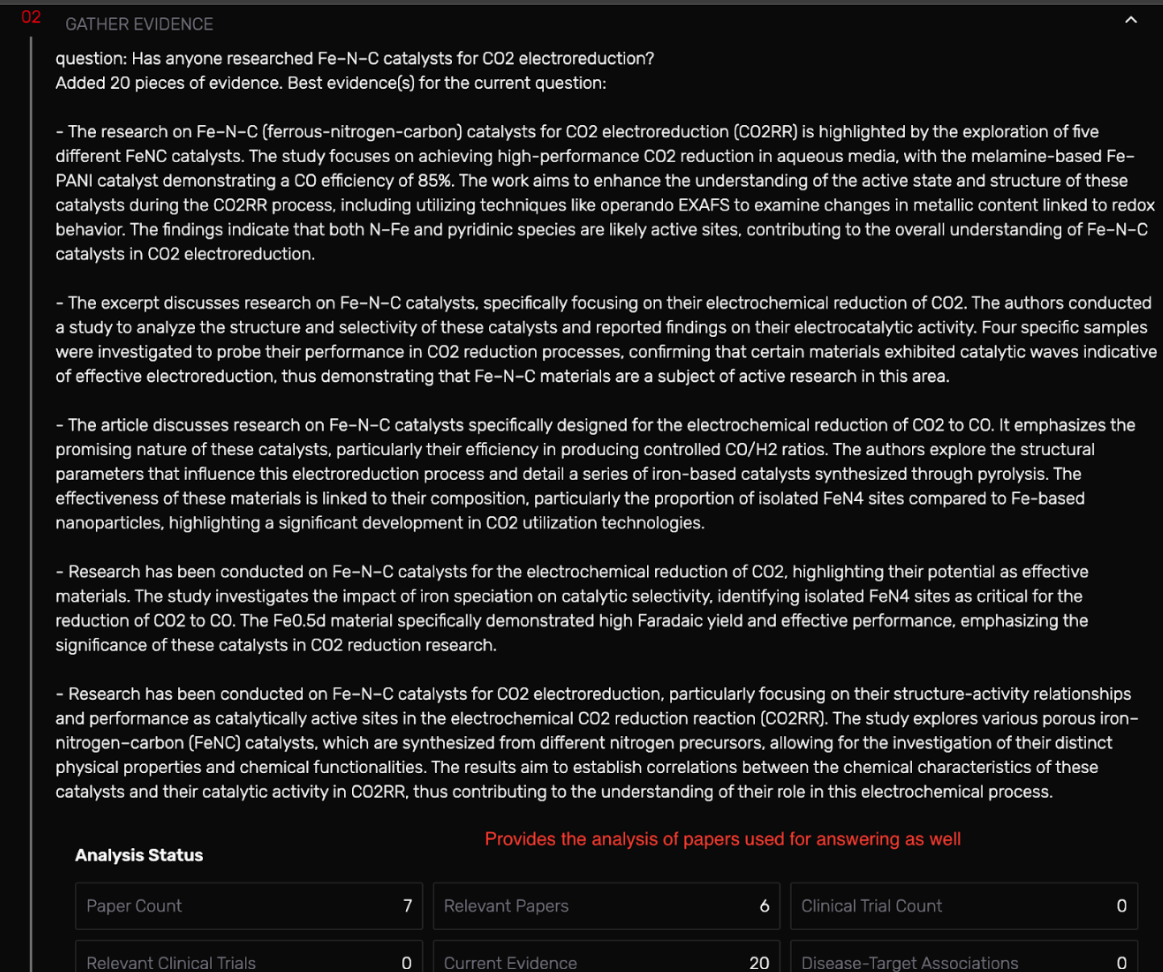
- Collects the cited papers in evidence
- Gathered evidence with bullet points based on the literature provided
- Finally, gave a definitive “yes” or “no” answer based on the evidence
Despite this extensive “thinking stage”, it did confuse ALI with acute lung injury, so she had to run it again with a more specific ask.
Revised prompt:
Has anyone explored the role of alginate from Pseudomonas aeruginosa, in human airway models like air liquid interface (ALI) systems?
The response:
The tool landed many more scientific publications and after several minutes an error occurred, followed by a disappointing and anticlimactic “No relevant evidence available”.
▶ Nurdana’s Test (Click to expand)
The prompt:
Query: “Fe–N–C catalysts for CO₂ electroreduction.”
The response:
It took 2–4 minutes but once it worked, it grouped about 7 studies, showed structure–performance trends, and even provided a descriptive reasoning on the studies already being conducted in the field since 2015.
Pros & cons
| What worked | What didn’t |
|---|---|
|
|
Microbiologist’s takeaway
For Nurdana, this type of tool offers a glimpse into the future of research automation. It goes beyond summarizing existing studies to connect results and even suggest what might be worth testing next. She described the experience as “more like brainstorming with a knowledgeable colleague than using a search tool.”
Antonella’s verdict was more measured. She found that the tool works reliably overall, appreciates that the links it provides are functional and lead to legitimate data, and noted that the “evidence” section aligns well with the cited papers. She found the interface easy to use, though not always fast, something she attributed either to computational load or heavy server traffic.
However, both testers agreed that the tool still lacks full transparency in its reasoning process and requires further refinement. As a result, they felt it may not yet be ready for everyday research use.
SciSpace
How it works
SciSpace (formerly Typeset) is an integrated AI workspace for researchers that brings together multiple tools to help you from the first literature search to refining the final manuscript. It has 130+ agents that assist, including tools for literature search, “Chat with PDF,” AI writer, citation generator, and data extraction, into a single platform. It also has a tool to detect AI generated text.
Testing
▶ Nurdana’s Test (Click to expand)
The prompt:
I uploaded a 2020 paper on Cryptococcus curvatus lipid extraction and asked SciSpace to summarize its methodology and outcomes.
The response:
It returned a clear breakdown of the nitrogen source, culture medium, extraction solvent, and analytical technique (GC-FAME), along with a note about the small experimental volume.
Follow-up prompt:
Using the dashboard, I could then switch directly to “Write a Report.”
The response:
The tool successfully drafted a short methods summary, automatically formatted and referenced.
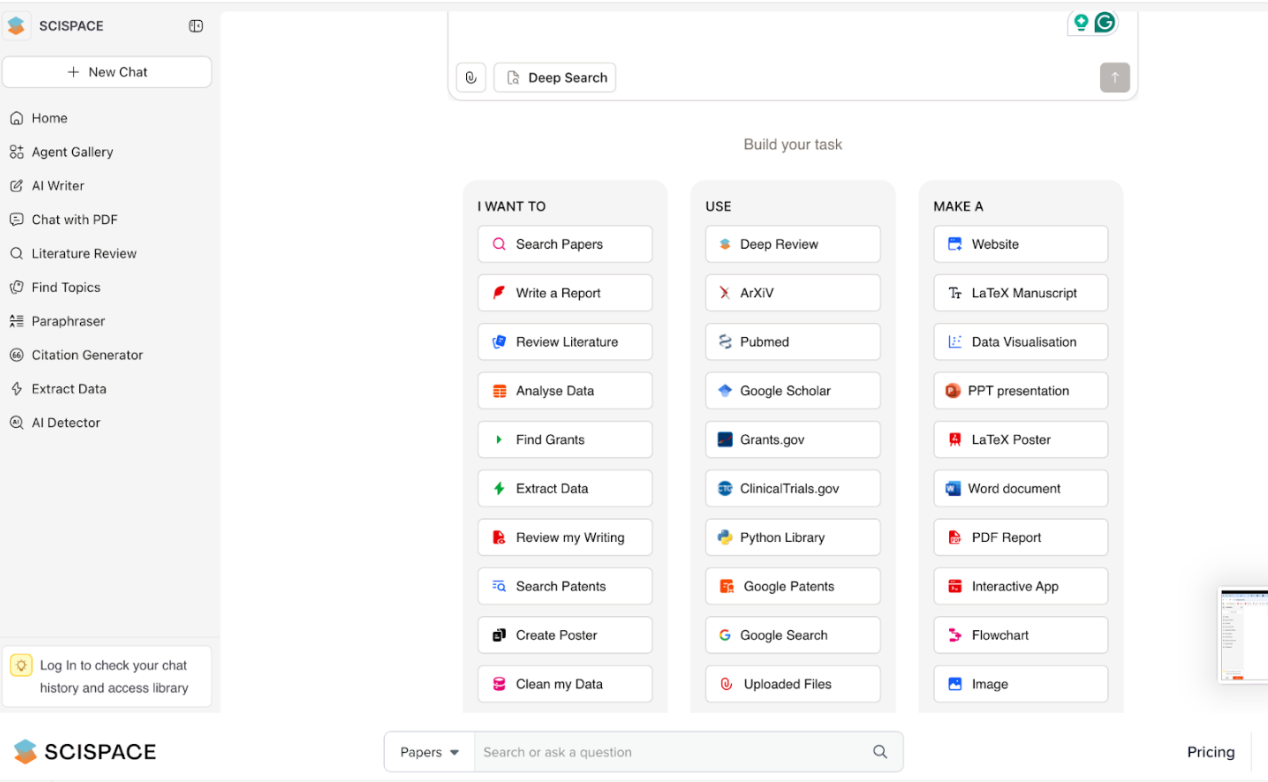
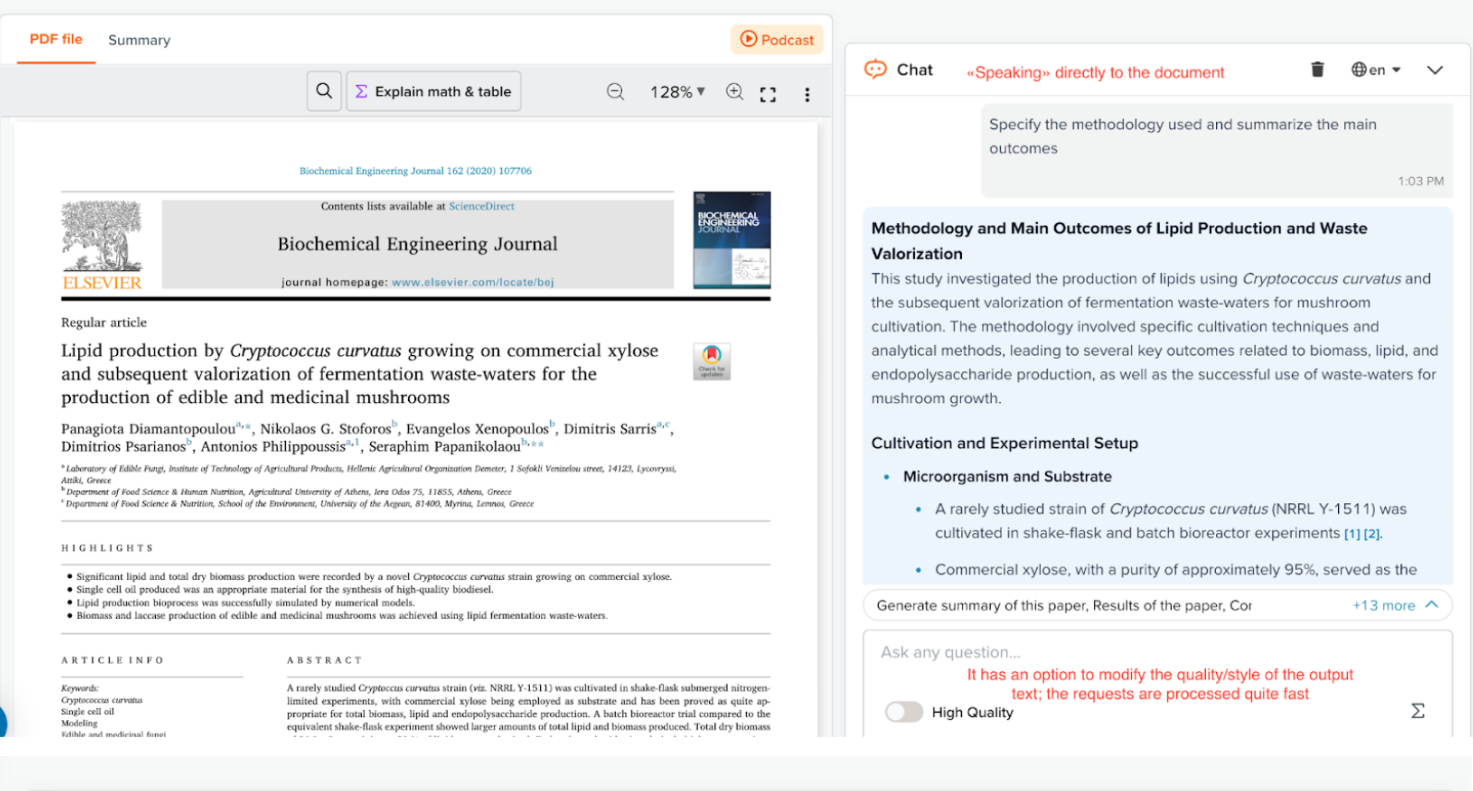

▶ Antonella’s Test (Click to expand)
The prompt:
I’m interested in the analysis done in this paper:
https://www.pnas.org/doi/10.1073/pnas.2221542120.
Can you describe the overview of what they want to achieve?
(I’m very familiar with the article, so I will be able to spot if it made mistakes.)
The response:
The first time, unfortunately, the program didn’t return anything valuable.
Attempt 2: Antonella tried the same test again at a later date. This time, she received results.
TL;DR
The authors developed and validated two quantitative, generalizable approaches to systematically improve laboratory models of microbial behavior by comparing gene expression in models versus natural environments, using Pseudomonas aeruginosa in cystic fibrosis lungs as proof of concept [1].
Key Takeaways
- Problem addressed: Laboratory models often fail to accurately represent microbial physiology in natural environments, but systematic approaches to improve them are lacking.
- Novel framework: The study quantifies model accuracy by comparing gene expression patterns between laboratory models and the natural environment (CF sputum).
- Two improvement strategies: (1) Combining complementary models with different strengths, and (2) mining transcriptomic data to identify missing environmental cues.
- Concrete outcomes: Achieved ~90% accuracy in gene expression by adding zinc limitation to synthetic CF sputum medium (SCFM2).
- Broader impact: These approaches are generalizable to any microbial system, potentially transforming how researchers develop and validate laboratory models.
Structured Overview
Research Motivation and Gap
The authors begin by acknowledging that laboratory models are fundamental to microbiology research, serving as tractable systems to study microbial cell biology and investigate inaccessible clinical/environmental ecosystems. However, they identify a critical problem: despite heavy reliance on these models, there is no systematic, quantitative framework for improving their relevance to natural environments [1].
Traditional model development has been largely empirical and intuitive rather than data-driven. While researchers have made advances (e.g., humanized mice, fabricated microbial ecosystems), the field lacks rational, generalizable approaches to assess and enhance model accuracy.
Core Objective
The primary goal of this study is to develop and validate generalizable, quantitative frameworks for improving microbiological model systems [1]. The authors use Pseudomonas aeruginosa infection in cystic fibrosis (CF) lungs as their proof-of-concept system because:
- P. aeruginosa is a major pathogen in CF patients
- CF sputum samples provide access to the “natural environment” (in vivo gene expression)
- Multiple existing laboratory models exist for comparison
Methodological Framework
The authors build upon their previously developed framework that assesses model accuracy by:
- Quantifying how closely each gene is expressed in the natural environment versus various laboratory models
- Defining model accuracy as the percentage of genes that show similar expression patterns between the natural environment and the model [1]
This quantitative metric allows objective comparison and improvement of models.
Two Improvement Strategies
Strategy 1: Model Combination
The authors identified that different laboratory models accurately recapitulate different subsets of genes:
- SCFM2 (synthetic CF sputum medium): An in vitro chemical model
- Epithelial cell model: A host-cell-based infection model
By combining these two models into a hybrid “epithelial cell-SCFM2 model,” they improved the accuracy of over 500 genes [1]. This approach leverages the complementary strengths of different model systems.
Strategy 2: Data Mining for Missing Environmental Cues
To improve accuracy for specific gene sets, the authors:
- Mined publicly available transcriptome datasets
- Identified zinc limitation as an environmental cue present in CF lungs but absent in SCFM2
- Induced zinc limitation in SCFM2, which resulted in accurate expression of ~90% of P. aeruginosa genes [1]
This demonstrates how systematic analysis of transcriptomic data can reveal specific missing conditions that, when added, dramatically improve model fidelity.
Broader Significance
The authors emphasize that these approaches are not limited to P. aeruginosa or CF infections. The frameworks they developed are generalizable and can be applied to:
- Any microbial system of interest
- Various natural environments (clinical, environmental, industrial)
- Different types of microbes and microbial communities
The study represents a shift from intuitive, trial-and-error model development to systematic, data-driven model optimization [1].
Ultimate Vision
The authors suggest these rationalized frameworks have the potential to reshape how we understand the role of microbes across ecosystems by ensuring that laboratory findings more accurately reflect natural microbial physiology [1].
References
[1] Article content from:
https://www.pnas.org/doi/10.1073/pnas.2221542120
(Accessed: November 26, 2025)
Feedback:
I then went an ask, do you understand how accuracy score (AS2) are being calculated? They have a model accuracy framework based on a direct comparison of the expression of microbial genes in the natural environment (in vivo lungs) and their laboratory model. Would you be able to calculate the accuracy score (AS2) of my infection model if I give you the input of RNA-seq read count raw data?
Read webpage
Searching for the full text PDF to access the methods section
I can see the scraped content only contains the abstract and significance statement. Let me try to access the full article with methods details about the AS2 calculation.
Browsing
https://www.pnas.org/doi/10.1073/pnas.2221542120
Accessing the full PDF to find the AS2 calculation methodology
Clicked
Agent is on a coffee break
You have exhausted your free credits. To continue using SciSpace Agent, upgrade to a higher plan.
Upgrade Now
Unfortunately, as Antonella asked additional questions, the available credits were exhausted. The tool appears to allocate a limited number of credits per day, which decrease with each query. So, when she repeated the same prompts, the first task (manuscript summary), it was completed quickly, but the second used up the remaining credits and returned no response.
Pros & cons
| What worked | What didn’t |
|---|---|
|
|
Microbiologist’s takeaway
The reviews were mixed. While Nurdana appreciated the results SciSpace provided and was particularly impressed by the “Chat with PDF” feature, Antonella, at first, struggled to get meaningful output due to paywall restrictions and system crashes. While Antonella’s second test round went better, she didn’t particularly feel like there was much additional value. She also expressed that the abundance of available tools and little explanation seemed overwhelming and difficult to navigate.
ChatGPT
How it works
ChatGPT is a large language model that can generate, summarize, and explain text across topics. While it’s not a scientific search engine, it can assist researchers in brainstorming ideas, summarizing concepts, explaining methods, or structuring reports when provided with sufficient context or references.
Testing
During the first attempt, when the apps crashed Antonella, she decided to run the same tests on ChatGPT. This is what she found.
▶ Antonella’s Test (Click to expand)
The prompt:
I’m interested in the analysis done in this paper
https://www.pnas.org/doi/10.1073/pnas.2221542120
Can you describe the overview of what they want to achieve?
The response:
The tool quickly generated accurate, logically structured explanations that included background, objectives, methods, and results summaries.
Follow-up prompt:
Do you understand how accuracy scores (AS2) are calculated in this RNA-seq model?
The response:
When asked about the AS2 calculation, it outlined step-by-step instructions on how to provide the data for analysis, making the process clear and approachable even for complex computational questions.
Pros & cons
| What worked | What didn’t |
|---|---|
|
|
Microbiologist’s takeaway
One of the testers found ChatGPT to be the most reliable tool overall, especially when other platforms were limited by paywalls or credit systems. They reported that it “gave accurate answers very fast, with detailed explanations on methods and even a comparison table,” noting that it was often the easiest and most dependable option in practice.
Limitations & use
| Tool | Good for | Limitations | Speed | Free use |
| Perplexity | Quick topic exploration with citations | Lacks experimental depth | ⚡ Fast | ✅ Basic use |
| Elicit | Structured data extraction, evidence tables | Slow, limited coverage | Moderate | ✅ Basic use |
| FutureHouse | Pattern recognition, research gaps | Unstable, chemistry-biased | Slow | ⚠️ Limited beta |
| SciSpace | End-to-end research management | Some features paid; simplified summaries | ⚡ Fast | ⚠️ Daily limit |
| ChatGPT | Brainstorming, rewriting, explaining concepts | Not built for research; may hallucinate sources | ⚡ Fast | ✅ Basic use |
Using AI responsibly:
- Start small: Try one use case first, e.g., use Perplexity just for literature scoping
- Verify every claim: Always check the original papers before citing
- Prompt precisely: Specific, detailed prompts reduce error and waste less computation.
- Stay transparent: Record which AI tools were used when writing or reviewing papers.
- Use AI for acceleration, not automation: The value lies in freeing time to think, not replacing scientific reasoning.
- Make sure you follow journal-specific editorial requirements: Every journal has their own editorial guidelines and therefore their own policy on AI and its allowed use. Please make sure your final submission follows these guidelines to improve your chances of publication.
Final thoughts from the researchers
Like most emerging technologies, the usefulness of AI tools depends greatly on the user. One of our testers found that most platforms were limited by paywalls, credit restrictions, or crashes, and preferred the reliability and responsiveness of ChatGPT. The other saw tremendous potential in the newer tools, noting how they could help connect ideas, accelerate literature reviews, and reveal research gaps that might otherwise go unnoticed.
As one researcher put it:
“No single tool replaces a researcher’s judgment or expertise, but together, they reshape how efficiently we can move from question to insight. The real advantage lies not in automation but in acceleration — freeing time from mechanical searching and formatting to focus on thinking, connecting, and creating new ideas. AI is not the scientist; it’s the lab assistant.”
Perhaps that’s the best way to view AI in microbiology research, not as a replacement for scientific reasoning, but as a companion that, when used responsibly, helps scientists spend more time on what truly matters.

