Vibrio cholerae is a human pathogen found in contaminated seafood or water. Since infections of this pathogen are on the rise due to climate change, we need to better understand its virulence regulators. The short review “Small regulatory RNAs in Vibrio cholerae” in microLife explores one such transcriptional regulation mechanism which is crucial for the pathogenicity process, as Rabea Ghandour presents for the #FEMSmicroBlog. #FascinatingMicrobes
About the pathogen Vibrio cholerae
Vibrio cholerae is a human pathogen causing intestinal infections that can be fatal within hours. Once ingested, Vibrio cholerae secretes various toxins that attack the host leading to severe dehydration.
Several cholera outbreaks have been reported in recent years and more are anticipated in the future due to natural disasters accompanying climate change. Thus, a better understanding of Vibrio cholerae’s infection process and pathogenesis is required.
As part of the infection cycle, Vibrio cholerae survives in and on sea animals before being ingested with contaminated food or water. Vibrio cholerae has developed several strategies to adapt to its environment, and employs several transcriptional and post-transcriptional regulators to optimize its gene expression in response to environmental cues.
Small regulatory RNAs in Vibrio cholerae
In the last two decades, numerous studies described small regulatory RNAs as crucial regulators of bacterial gene expression. Small regulatory RNAs form base pairs with their target mRNAs and modulate their secondary structure. This interaction changes the stability of the target mRNA, triggering or inhibiting its decay and impacting its translation rates.
In most cases, small regulatory RNAs function in concert with RNA-binding proteins, such as the highly conserved Hfq protein. In Vibrio cholerae, Hfq mediates interactions of small regulatory RNAs with plenty of mRNA targets as shown by global RNA-RNA interaction studies.
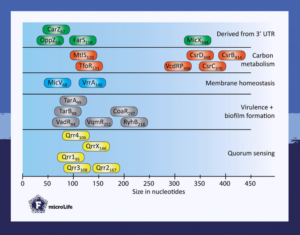
The short review “Small regulatory RNAs in Vibrio cholerae” in microLife explores how these regulators control multiple cellular processes in Vibrio cholerae ranging from intercellular communication, such as quorum sensing, to biofilm formation, bacterial warfare, virulence, carbon metabolism, and membrane homeostasis.
Lifestyle decisions based on small regulatory RNAs
Initially, small regulatory RNAs in Vibrio cholerae were found to be connected to quorum sensing. Now it is clear that this bacterium possesses a large repertoire of small regulatory RNAs integrating a wide range of sensory inputs.
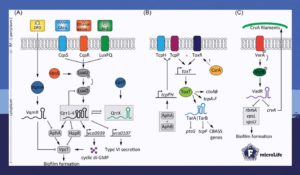
For example, upon sensing low levels of autoinducers on the bacterial surface, the small regulatory RNAs Qrr1-4 are activated. These regulate cyclic di-GMP concentration, biofilm formation, and activity of the type 6 secretion system.
Furthermore, Qrr1-4 are negatively controlled by the RNA sponge QrrX. This small regulatory RNA binds to Qrr1-4 through extensive base-pairing which triggers degradation of the RNA duplexes. While the activation mechanism of QrrX is still not clear, this interaction modulates several lifestyle choices in Vibrio cholerae.
Another small regulatory RNA controlling lifestyle decisions in Vibrio cholerae is VadR. In contrast to QrrX, this small regulatory RNA does not depend on quorum sensing, and yet inhibits the expression of several genes involved in biofilm matrix production.
Additionally, VadR also downregulates the translation of the crvA mRNA. This gene encodes for a periplasmic protein involved in giving Vibrio cholerae its characteristic curve shape. Hence, upon sensing cell-wall damaging antibiotics and mechanical stress, VadR coordinates the bacterium’s cell shape, antibiotic tolerance, and biofilm formation.
This review gives a comprehensive overview of different aspects of control of gene expression by small regulatory RNAs in Vibrio cholerae, promoting a richer understanding of the physiology of this pathogenic bacterium.
- Read the review “Small regulatory RNAs in Vibrio cholerae” in microLife by Ghandour and Papenfort (2023).
Rabea Ghandour is a postdoc in the Institute of Microbiology at the Friedrich Schiller University Jena in the group of Kai Papenfort. She completed her PhD studies at the Max Plank Institute of Molecular Plant Physiology where she worked on bacterial translational feedback regulation. Rabea currently works on small regulatory RNAs and RNA-binding proteins in the human pathogen Vibrio cholerae. She has a lot of interest in RNA biology and post-transcriptional regulation.
About this blog section
The section #FascinatingMicrobes for the #FEMSmicroBlog explains the science behind a paper and highlights the significance and broader context of a recent finding. One of the main goals is to share the fascinating spectrum of microbes across all fields of microbiology.
| Do you want to be a guest contributor? |
| The #FEMSmicroBlog welcomes external bloggers, writers and SciComm enthusiasts. Get in touch if you want to share your idea for a blog entry with us! |

