Bacteria barely ever live in isolation, but rather in multispecies communities. Within these communities, they interact, thus impacting the growth, shape, and functions of the community. The study “Inferring bacterial interspecific interactions from microcolony growth expansion“ in microLife investigates the kinetic variations in bacterial mono- and co-cultures. Tania Miguel Trabajo, Isaline Guex and Jan Roelof van der Meer discuss new insights into bacterial interactions, from local to community-level dynamics. #FascinatingMicrobes
Bacterial interactions in microbial communities
Within microbial communities, different types of interactions occur. These include, for instance, competing for growth substrates as well as inhibiting through or depending on excreted chemical compounds. Aggression, such as physical stabbing and sucking nutrients from cells through macromolecular structures, is also common among interacting bacteria.
However, until now, no consensus on how to measure interspecies interactions exists. Established methods are either indirect or aggregate global behaviour. Often, multiple species are grown in mixed liquid cultures where spatial context is lost.
Even though bacterial cells occupy physical space, they have only a limited number of direct neighbours with whom they interact. That’s why the study “Inferring bacterial interspecific interactions from microcolony growth expansion“ in microLife aimed to develop a method to observe these emerging interactions at the level of individual cells. The study further quantified interactions to create mathematical growth models.
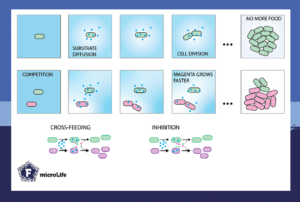
An experimental setup to observe interacting bacteria
The study settled for time-lapse microscopy to observe how sparsely seeded cells divide on surfaces and form small microcolonies. By restricting the total available carbon substrate, colonies would remain sufficiently small and establish only sporadic contacts between them. Yet, this setup would still allow the observation of growth kinetic parameters such as lag, exponential, and stationary phases.
By comparing microcolony growth of pure cultures to that in mixtures and on different substrates, one could deduce whether and what type of interactions emerge. To describe and understand the kinetics of the observed microcolony growth, a comparative mathematical model was made. This was based on in silico cells that grow and divide into microcolonies as real bacteria would do. The model revealed the effects of interactions by removing or adding mathematical factors to the in silico cells.
Yet, this experimental setup came with several challenges: The microscopy experiments generated large amounts of data for which colony measurements needed to be automated, even from imperfect images. Furthermore, the amount of nutrients and the density of seeded cells needed to be calibrated to limit overgrowth. Finally, during bacterial growth, nutrient gradients arose in the microscope chambers, leading to spatial heterogeneities.
Learning from bacterial interactions
In the simplest experiment, the two bacterial species Pseudomonas putida and Pseudomonas veronii grew together. As a result, both partners decreased in biomass and growth rates. These findings were likely due to the underlying competition between the two species for the substrate.
This was unexpected, since growth rates are supposed to be determined by substrate concentration, according to mathematical growth theory. However, the so-called cell-agent model postulates that the reduced growth rates may result from either excreting mutual inhibitory compounds or cross-feeding on metabolites as secondary substrates.
Previous research on cross-feeding showed that growing populations excrete a range of metabolites from the primary added carbon substrate. Other bacterial cells take these up and use them for growth but with lower energy and yield coefficients. The result of this cross-feeding is measurable as a reduced growth rate and, eventually, fewer cells produced from the added substrate.
The study further showed that bacterial interactions in co-cultures are constant across different cell neighbourhood areas. These interactions can even be quantified from the sizes of the microcolony areas of both partners.
Despite these constant interactions, the growth of 10-15 % of individual cells was affected due to cell-to-cell variability. Hence, one can observe both the average interaction behaviour and individual variation across different scales. At the same time, simultaneous measurements of microcolony growth of different species can be used to both quantify the average type and strength of their interactions, as well as estimate their individual variation.
Measuring at a local scale may permit to infer growth interactions among more than two species at the same time, when their colonies intermingle in various configurations. In summary, several components are important to understand how communities of different bacterial species form and maintain.
- Read the article “Inferring Bacterial Interspecific Interactions from Microcolony Growth Expansion” by Trabajo et al. in microLife (2024).
Tania Miguel Trabajo recently graduated with a PhD from the University of Lausanne. She now works for Sophia Genetics.
Isaline Guex gained her PhD in Mathematics from the University of Fribourg in 2024 and is now working as a postdoc in the group.
Jan Roelof van der Meer is Professor for Environmental and Evolutionary Microbiology at the Department of Fundamental Microbiology at the University of Lausanne. He is also the current Director of the Swiss network in microbiomes research, named NCCR Microbiomes.
About this blog section
The section #FascinatingMicrobes for the #FEMSmicroBlog explains the science behind a paper and highlights the significance and broader context of a recent finding. One of the main goals is to share the fascinating spectrum of microbes across all fields of microbiology.
| Do you want to be a guest contributor? |
| The #FEMSmicroBlog welcomes external bloggers, writers and SciComm enthusiasts. Get in touch if you want to share your idea for a blog entry with us! |
