The urinary tract is a challenging environment for bacterial pathogens. Yet, uropathogenic Escherichia coli manages to withstand urine flow and reach the kidneys where they cause disease. The article “Dynamic single cell analysis in a proximal-tubule-on-chip reveals heterogeneous epithelial colonization strategies of uropathogenic Escherichia coli under shear stress” in FEMS Microbes investigates how this pathogen tolerates these conditions and colonizes the kidneys. Haris Antypas explains in a #BehindThePaper interview the importance of these findings.
Can you explain the importance of the paper for a general audience?
When uropathogenic E. coli enter the urinary tract from the urethra and reach the kidneys, they can cause an infection called pyelonephritis. If left untreated, this infection can damage the kidneys and lead to a serious condition called sepsis.
During the infection process, the bacteria are facing the flow of urine along the way. And it is still not understood how they can withstand this flow on their way to the kidneys.
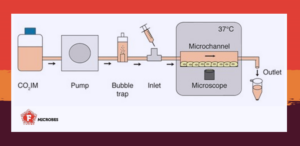
To shed light on this question, we used a method called organ-on-a-chip, which involves creating miniaturized artificial organs, like the kidneys. This approach allowed us to study how diseases develop in a more realistic environment than traditional lab methods.
We found that uropathogenic E. coli has a protein called PapG that enables them to stick to kidney cells for longer periods of time. With this adhesion, the pathogen buys time to rapidly multiply and form groups called microcolonies.
Within these microcolonies, the bacteria can stick together and overcome the force of the fluid flow. We also found that the protein FimH plays a role in forming these microcolonies by helping bacteria interact with each other.
Overall, this research could help develop treatments for urinary tract infections by targeting these bacterial proteins and preventing them from interacting with kidney cells and each other. It also shows how the organ-on-a-chip method can be used to study infection without using animals.
How does high-resolution real-time analysis using new technologies help advance the microbiology field?
Studying bacteria at a single-cell level in real time can give us a better understanding of microbial communities and their phenotypic heterogeneity during infection. Cell culture and animal models have provided valuable data to help us understand host-pathogen interactions. However, both approaches tend to focus on the infection endpoint, which may not show how and at which stage virulence factors contributed to the infection.
The organ-on-a-chip technology allowed us to mimic the renal proximal tubule and expose bacteria to the shear stress they face in vivo when trying to colonize the renal epithelium. This workflow provided a controlled environment to follow single bacteria with microscopy from initial adhesion to microcolony formation and widespread colonization of renal epithelial cells.
Traditionally, the study of virulence factors, such as fimbriae, has been conducted in endpoint assays. However, our real-time analysis provided a more comprehensive understanding of the adhesion process of uropathogenic E. coli under fluid flow and the role of PapG and FimH adhesins in colonization.
Why did you choose to dive into the topic of this paper? What fascinates you about the stress adaptation of uropathogenic E. coli?
The possibility of obtaining a fresh perspective on this pathogen’s ability to infect renal cells through an unconventional approach initially captivated me. It was after attending a presentation by the Richter-Dahlfors group, who used an intravital model in rats to track kidney infection in real time.
As an MSc student at the Karolinska Institutet, the prospect of studying host-pathogen interactions in real-time was unprecedented to me. During an interview with Prof. Agneta Richter-Dahlfors, she revealed that she aimed to simulate pyelonephritis on a chip, which turned into my primary research project as a PhD candidate.
While working on the project, I became increasingly fascinated by the adaptability of uropathogenic E. coli in a shear-stress environment. I was amazed by the complexity of virulence and the way that timing plays a crucial role in the progression of the infection.
It was intriguing to discover how adhesion factors play different parts at different stages of infection and how they influenced the outcome of the infection. Overall, my work on uropathogenic E. coli virulence was a truly engaging and enriching experience that certainly encouraged me to pursue research in the field of bacterial pathogenesis.
Why did you choose the transparent peer-review process for your publication?
I believe that a transparent peer-review process encourages reviewers to be more accountable for their comments and provide better feedback. This can help in eliminating biases from the reviewers’ side and lead to fairer peer reviewing.
At the same time, good peer-reviewing takes time and effort and I think it is great to be publicly acknowledged and get credit for peer-reviewing an article. Finally, I think that being able to read the exchange between the reviewer and the authors is a great way for junior scientists to understand what experiments are required to support their research findings and draw sound conclusions.
- Read the paper “Dynamic single cell analysis in a proximal-tubule-on-chip reveals heterogeneous epithelial colonization strategies of uropathogenic Escherichia coli under shear stress” by Antypas et al. (2023) in FEMS Microbes.

Dr. Haris Antypas is a senior research fellow in the Kline Lab at the Singapore Centre for Environmental Life Sciences Engineering (SCELSE). He specializes in infective endocarditis and catheter-associated urinary tract infections. He is studying how bacteria establish biofilms in our body and how our immune system fights off biofilms using animal models and microfluidics, with the long-term goal to identify new therapeutic targets. Haris earned his PhD from Karolinska Institute, where he developed tools for studying and diagnosing biofilm infections. The development of a method to diagnose biofilm-related urinary tract infections during his PhD earned him a patent and a postdoctoral fellowship from the European Institute of Technology. At SCELSE, he’s received the Wallenberg-NTU Postdoctoral Fellowship, SCELSE Seed Funding, and Young Individual Research Grant from NMRC, all of which help support his research on the role of neutrophils in infective endocarditis biofilms.
About this blog section
#BehindThePaper posts on the #FEMSmicroBlog aim to bring science closer to different audiences and to tell more about the scientific or personal journey to come to the results.
| Do you want to be a guest contributor? |
| The #FEMSmicroBlog welcomes external bloggers, writers and SciComm enthusiasts. Get in touch if you want to share your idea for a blog entry with us! |
