The increasing problem of antibiotic-resistant bacteria requires us to develop more sensitive and accurate detection methods. Quantitative PCR and metagenomics are normally used to quantify the presence of certain genes within a microbial community. The study “Quantitative PCR versus metagenomics for monitoring antibiotic resistance genes: balancing high sensitivity and broad coverage” in FEMS Microbes uses these two methods to screen animal faecal, wastewater and water samples as explained by Célia Manaia in a #BehindThePaper interview.
Can you explain the importance of the paper for a general audience?
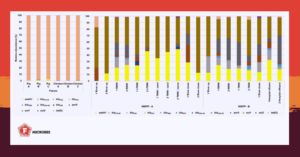
Our paper is about quantitative methods that can be used to monitor antibiotic resistance in animal faecal material and in wastewater and water. This monitoring is important to inform about the occurrence of antibiotic resistance and if antimicrobial resistance genes can disseminate from there.
How do quantitative methods like quantitative PCR and metagenomics help advance the microbiology field?
Metagenomics is a fabulous methodology to get an overview of microbial communities, as if we were seeing a landscape through a drone flying over a dense forest. With this technique it is possible to have comprehensive insights into the vast majority of the microbial species that inhabit a given environment. We see the forest, which is the whole community of microorganisms as well as their genes.
With quantitative PCR we may look for the small bush that would not be easily detected with the panoramic overview. In this case, we are looking for a specific bush, what it looks like and how big it is. When looking at a specific player, we don’t care about the forest as a whole.
Quantitative methods are relevant in areas such as the dissemination of antibiotic resistance. Here, the quantity is important as it can be indicative of risks or the need for control measures. These studies can also simply infer about possible antibiotic resistance reservoirs.
Why did you decide to dive into the topic of this paper? What fascinates you about bacterial antimicrobial resistance?
Some antibiotic-resistant bacteria are fascinating examples of how living organisms can succeed at adaptation. This mechanism allows them to grow faster than others, survive adverse conditions, share genetic information or provide shelter to their neighbours to strengthen the community. In short, they can adapt to different types of challenge.
However, this fascinating mechanism is also the reason why antibiotic-resistant bacteria are an increasing problem for human health. That’s why they need to be urgently tackled through solutions that come from multiple areas of knowledge and intervention.
Why did you choose the transparent peer-review process for your publication?
The peer-review process can be a stimulating and interesting exchange of opinions and points of view. This includes getting the perspective of experts that are outside the core topic that we are reporting.
As an author, whenever possible, I regard the peer-review process as way of learning – a point of view that I never thought about, an information that I did not know, an incentive to reinterpret data… So, when that is an option, why not make this process transparent and open?
- Read the paper “Quantitative PCR versus metagenomics for monitoring antibiotic resistance genes: balancing high sensitivity and broad coverage” by Ferreira et al. (2023) in FEMS Microbes.
 Célia Manaia is an associate professor at Universidade Católica Portuguesa, teaching microbiology and genetics. Her research interests focus on bacterial taxonomy, evolution and ecology, with a special interest in human-environment interfaces and in the environmental dispersion of antibiotic-resistant bacteria. She has integrated international consortia and worked with business and public entities, interested in the evaluation and development of public policies. As part of her activity as a microbiologist, she is currently vice-president of the Portuguese Society of Microbiology, a FEMS member society.
Célia Manaia is an associate professor at Universidade Católica Portuguesa, teaching microbiology and genetics. Her research interests focus on bacterial taxonomy, evolution and ecology, with a special interest in human-environment interfaces and in the environmental dispersion of antibiotic-resistant bacteria. She has integrated international consortia and worked with business and public entities, interested in the evaluation and development of public policies. As part of her activity as a microbiologist, she is currently vice-president of the Portuguese Society of Microbiology, a FEMS member society.
About this blog section
#BehindThePaper posts on the #FEMSmicroBlog aim to bring science closer to different audiences and to tell more about the scientific or personal journey to come to the results.
| Do you want to be a guest contributor? |
| The #FEMSmicroBlog welcomes external bloggers, writers and SciComm enthusiasts. Get in touch if you want to share your idea for a blog entry with us! |
