Bacterial infections are again becoming increasingly difficult to treat due to the rise of resistance to antibiotics. One bacterial signaling mechanism could be an antimicrobial target, as presented in “The role of site-2-proteases in bacteria: A review on physiology, virulence, and therapeutic potential” in microLife. In this #FEMSmicroBlog, Sofie S. Kristensen explains how bacterial pathogens use site-2-proteases for adaptation and virulence, which may render these enzymes potential targets for antimicrobial drugs. #FascinatingMicrobes
Bacteria adapt to their environment
To survive, bacteria need to sense and respond to changes in their extracellular environment. Several Gram-positive and Gram-negative species rely on a signal transduction mechanism known as “regulated intramembrane proteolysis”.
Regulated intramembrane proteolysis is a conserved signaling mechanism generally involving two membrane-bound proteases: A site-1-protease and a site-2-protease. These two proteases subsequently degrade a substrate, for example, an anti-sigma factor. The degradation of the substrate results in the activation or deactivation of target gene expression and thereby an adaptive response (Figure 1).
The short review “The role of site-2-proteases in bacteria: a review on physiology, virulence and therapeutic potential” in microLife presents how regulated intramembrane proteolysis is involved in different cellular processes. It describes site-2-proteases as master regulators of this signaling mechanism, and as valuable antimicrobial targets in bacterial pathogens.
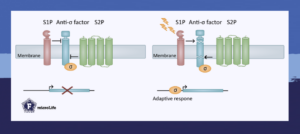
Pathogenic bacteria use regulated intramembrane proteolysis during infection
Pathogenic bacteria are reliant on a quick adaptation to the environment. During infection, pathogenic bacteria face challenges, such as limited access to iron, exposure to antimicrobials, or chemical attacks from the host immune system. To combat these challenges, several pathogens rely on regulated intramembrane proteolysis.
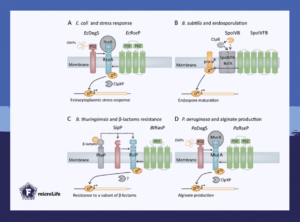
The human pathogen Pseudomonas aeruginosa relies on a site-2-protease to release multiple sigma factors and activate its iron acquisition systems. Similarly, several Gram-positive bacteria, including enterococci, different Bacillus species, and Clostridium difficile, depend on regulated intramembrane proteolysis to activate genes to resist lysozyme and β-lactam antibiotics. Mutations in site-2-proteases severely reduce the pathogen’s ability to respond to changes in the environment, often resulting in reduced infection rates and survival.
While several pathogens rely on regulated intramembrane proteolysis to respond to changes in the host environment, others use this mechanism to directly activate virulence factors. In Mycobacterium tuberculosis, removing the site-2-protease results in loss of virulence traits as well as reduced growth and persistence in animal models.
In P. aeruginosa, a site-2-protease is linked to mucoid production to protect the pathogen from host attacks. Removal of site-2-proteases in other human pathogens, like Staphylococcus aureus, Enterococcus faecalis, or Enterococcus faecium, reduces virulence. As site-2-proteases play key roles in the adaptation and virulence of several human pathogens, they represent promising antimicrobial targets.
Site-2-proteases as antimicrobial targets
Site-2-proteases act as molecular docking stations for specific antimicrobial peptides known as bacteriocins produced by several Gram-positive bacteria. The antimicrobial activity of bacteriocins depends thus on the interaction with the site-2-protease in the prey bacterium.
While the bacterium may respond with mutations in the site-2-protease to become resistant to the bacteriocin, this comes at a high cost as the protease is essential for adaptation and virulence. Spontaneous bacteriocin-resistant mutants with modified site-2-proteases show decreased abilities to adapt to environmental changes and reduced survival levels.
In this sense, site-2-proteases represent the Achilles heel of bacteria: the protein renders the bacteria susceptible to antimicrobial drugs, while changes in the protein reduce their ability to cause infection. Since site-2-proteases are conserved in several important human pathogens, they represent promising targets for antimicrobial drugs.
- Read the article “The role of site-2-proteases in bacteria: A review on physiology, virulence and therapeutic potential” in microLife by Kristensen et al. (2023)

Sofie S. Kristensen holds a MSc in biomedical science from the University of Western Australia and recently earned her PhD from the Norwegian University of Life Science. In her work, Sofie focuses on the interaction mechanism between the site-2-protease RseP and small, antimicrobial peptides known as bacteriocins, with the long-term goal of developing bacteriocins as novel antimicrobials. Her research interest includes antimicrobial peptides, protein-protein interaction and antimicrobial resistance.
About this blog section
The section #FascinatingMicrobes for the #FEMSmicroBlog explains the science behind a paper and highlights the significance and broader context of a recent finding. One of the main goals is to share the fascinating spectrum of microbes across all fields of microbiology.
| Do you want to be a guest contributor? |
| The #FEMSmicroBlog welcomes external bloggers, writers and SciComm enthusiasts. Get in touch if you want to share your idea for a blog entry with us! |
