Tuberculosis has plagued humanity for millennia and remains the leading cause of bacterial disease-related deaths worldwide, with over 1.5 million deaths in 2022. Drug-resistant tuberculosis is the single largest component of the global antimicrobial resistance crisis with treatment being lengthy and complex and novel therapies currently lacking. The study “Mycobacterium tuberculosis infection triggers epigenetic changes that are enriched in a type I IFN signature” in microLife used epigenetic profiling to reveal novel genes and pathways that are dysregulated in tuberculosis infection. Katie Madden explains for the #FEMSmicroBlog how better understanding the complex nature of the tuberculosis immune response can help develop diagnostics and therapeutic targets. #FascinatingMicrobes
Mycobacterium tuberculosis reprograms host immune cells
Tuberculosis is caused by inhaling the bacterium Mycobacterium tuberculosis. This pathogen then infects immune cells in the lungs where it replicates and causes disease.
These host immune cells are specialized types of white blood cells called alveolar macrophages. Normally, they are very effective at eliminating bacteria that invade human airways. However, Mycobacterium tuberculosis reprograms these cells to impair key antibacterial responses so the pathogen can survive and persist upon infection.
Mycobacterium tuberculosis achieves this by modulating the packaging and organization of our genetic material – a field that biologists call epigenetics. These changes impact gene expression and thus protein production. Hence, after infection, the proteins and immune responses that the host cells have in their arsenal to combat Mycobacterium tuberculosis are weakened.
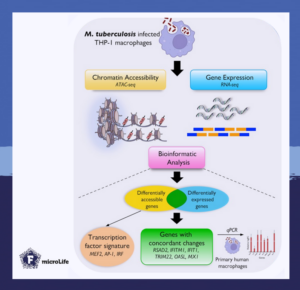
Regulation of gene expression is in part mediated by how accessible DNA strands are for transcription. If they are tightly packed, the transcription machinery cannot access them and these genes are not expressed, and vice versa.
The accessibility and expression of several genes involved in immune responses is modulated upon Mycobacterium tuberculosis infection. The paper presents the hypothesis that this phenomenon could be spread throughout the global genome and that Mycobacterium tuberculosis infection might affect hundreds of genes in macrophages.
Mycobacterium tuberculosis infection changes the human genome
The study “Mycobacterium tuberculosis infection triggers epigenetic changes that are enriched in a type I IFN signature” in microLife used next-generation genomic sequencing methods to profile the entire macrophage genome upon Mycobacterium tuberculosis infection.
Indeed, infection by Mycobacterium tuberculosis altered DNA accessibility and gene expression of hundreds of genes in macrophages. These changes are induced by specific human proteins. However, it is not clear whether the proteins are recruited by Mycobacterium tuberculosis or as part of the human immune response to bacterial infection.
Using bioinformatics analysis, the study further aimed to understand the functional relevance of this epigenetic dysregulation in Mycobacterium tuberculosis-infected macrophages. A significant proportion of the genes with changes in their DNA accessibility and gene expression are involved in ‘type I interferon (IFN)’ immune responses.
Interestingly, Type I IFN responses are potent, specialized immune responses that are effective at clearing viruses in humans. However, in the case of Mycobacterium tuberculosis infection, type I IFN immune responses could be detrimental to the host.
Upon Mycobacterium tuberculosis infection, changes to several type I IFN response genes in macrophages are mediated epigenetically. This could provide a new way to use these pathways as biomarkers or as targets for therapy.
This study uncovered multiple novel and potentially therapeutically relevant genes that have yet to be studied in the context of tuberculosis. With the rise of multi-drug-resistant tuberculosis and the lack of novel antibacterial therapies, developing potent therapy is imperative to combat the world’s deadliest bacterial pathogen.
- Read the article “Mycobacterium tuberculosis infection triggers epigenetic changes that are enriched in a type I IFN signature” by Madden et al. (2023).
- Read also “#FEMSmicroBlog: World Tuberculosis Day 2021″

Katrina Madden is a 4th year PhD candidate in the Department of Biochemistry, Microbiology, and Immunology at the University of Ottawa. She completed her undergraduate degree at the University of Waterloo and Master’s degree in Immunology and Global Health at Maynooth University in Ireland. Following her Master, Katie worked at Western university as a research/technical assistant with the HIV investigators group and the Imaging Pathogens for Knowledge Translation (ImPaKT) facility. Katie’s current research focuses on epigenetic regulation in tuberculosis. This research will define novel drug targets for adjunctive TB therapies and further elucidate host biology in Tuberculosis.
About this blog section
The section #FascinatingMicrobes for the #FEMSmicroBlog explains the science behind a paper and highlights the significance and broader context of a recent finding. One of the main goals is to share the fascinating spectrum of microbes across all fields of microbiology.
| Do you want to be a guest contributor? |
| The #FEMSmicroBlog welcomes external bloggers, writers and SciComm enthusiasts. Get in touch if you want to share your idea for a blog entry with us! |
