Small proteins are an emerging field of research while little is known about their functions – the least in archaea. Their minuscule protein size makes them a challenge not only to work with but also to discover. The study “Revealing the small proteome of Haloferax volcanii by combining ribosome profiling and small-protein optimized mass spectrometry” in microLife aimed to identify small proteins in a model archaeon. Lisa-Katharina Maier, Lydia Hadjeras and Jürgen Bartel explain for the #FEMSmicroBlog how two powerful approaches help shed light on archaeal physiology. #FascinatingMicrobes
The challenges of studying small microbial proteins
Over the past decade, modern genomics and transcriptomics have discovered a wealth of bacterial small genes that encode small proteins of <50 amino acids in length. Many common in silico and wet-lab approaches are biased against discovering these small proteins, leaving them often overlooked and thus poorly represented. Modern -omics-driven research setups rely on and are biased by the underlying annotation databases. Hence, small proteins must first be catalogued before investigating their full range of functions.
Due to their position within the tree of life, archaea are an interesting subject to study. Their replication and transcription machineries are eukarya-like, while their genome architecture and metabolism are similar to what is known from bacteria. Interestingly, while combining these bacterial and eukaryotic traits, archaea also have their own unique characteristics and regulatory features.
To better understand archaea and their physiology, one can focus on the functions of small proteins. Shedding light on how small proteins carry out their tasks will help identify common modes of action for small proteins across all forms of life. Or the answer may surprise us with unexpected pathways.
The study “Revealing the small proteome of Haloferax volcanii by combining ribosome profiling and small-protein optimized mass spectrometry” in microLife combines small protein-optimised mass spectrometry and ribosome profiling to identify annotated as well as novel small proteins in the model archaeon Haloferax volcanii.
The small proteome of Haloferax volcanii
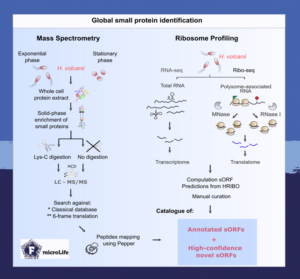
This study is the first multi-omics analysis of an archaeal small proteome. It captures low molecular weight proteins with mass spectrometry analysis and identified sites of ribosomal occupancy by ribosome profiling.
This analytical approach thus provided the so far most complete picture of the H. volcanii small proteome. The study identified 47 new small proteins and found that two-thirds of annotated small proteins are expressed under standard growth conditions.
Annotated small proteins were identified by both analyses using classical database searches for matching. However, the study took the analysis one step further by also subjecting the raw data of both mass spectrometry and ribosome profiling to annotation-independent bioinformatic examination.
For that, peptide spectra from mass spectrometry were matched to a 6-frame translation so that every possible genome-wide ORF was considered. Combining the two annotation-independent datasets revealed numerous sites outside the canonical annotation to be engaged in small protein biosynthesis.
Characterizing small archaeal proteins
The study further aimed to broadly characterise the expressed small proteins. Their small sizes often obstruct such studies since short protein sequences contain only a few functional domains. Additionally, small proteins tend to be less conserved.
Thus, it is of no surprise, that almost two-thirds of the expressed annotated small proteins are still uncharacterised. The remaining proteins have quite diverse functions and are ribosomal or cold-shock proteins that belong to the sec membrane transport system. Accordingly, the cellular localisations of the novel proteins vary greatly and they are predicted to reside in the cytosol, the membrane or to be secreted.
This study not only demonstrates that combining mass spectrometry and ribosome profiling is a powerful approach to investigate the translational activity, but also reminds us to revisit data independent of a priori assumptions. Even the proteome of a well-studied model organism may hold novel findings. Therefore, archaeal researchers could consider implementing combinatorial approaches to spotlight more small proteomes in their favorite model archaeon.
- Read the article “Revealing the small proteome of Haloferax volcanii by combining ribosome profiling and small-protein optimized mass spectrometry” by Hadjeras, Bartel, Maier et al. (2023).
About the authors of this blog
Lisa-Katharina Maier is a postdoctoral researcher in the Department of Molecular Biology and Biotechnology of Prokaryotes in the group of Prof. Anita Marchfelder at the University Ulm. Lisa has studied archaea throughout her scientific career with a focus on Haloferax volcanii‘s defense and transcriptional landscape. The research scope of the Marchfelder lab ranges from CRISPR-Cas system(s) over small RNAs and transcription up to now small proteins – short all things defence and RNA metabolism and with this publication entering the world of the small proteome.
Lydia Hadjeras is a passionate RNA biologist who completed her PhD in Molecular Biology at the University of Toulouse (France, Carpousis lab), where she worked on RNA degradosome localization in bacteria. As a postdoctoral researcher at the Institute of Molecular Infection Biology in Wuerzburg, (Germany, Sharma lab), Lydia continues to explore her passion for RNA biology and has extended her research interests to include the development of NGS-based technologies for transcriptome and translatome analyses in different prokaryotes. This published work on Haloferax volcanii small proteome raised from a fruitful collaboration between the Sharma lab, Marchfelder lab, Becher lab and Backofen lab, in the framework of a DFG-priority program “SPP 2002 – Small proteins in Prokaryotes, an unexplored world”.
Jürgen Bartel is a lab technician in the Group for Microbial Proteomics, led by Prof. Dörte Becher, at the University of Greifswald. Prior to joining this group, he studied metalloproteins and eukaryotic metalloproteomes which piqued his interest in developing innovative methods for MS-based proteomics. Currently, the Group for Microbial Proteomics is part of the “SPP 2002 – Small proteins in Prokaryotes, an unexplored world” and recently published an optimized protocol for the detection of small proteins, which was utilized in this study.
About this blog section
The section #FascinatingMicrobes for the #FEMSmicroBlog explains the science behind a paper and highlights the significance and broader context of a recent finding. One of the main goals is to share the fascinating spectrum of microbes across all fields of microbiology.
| Do you want to be a guest contributor? |
| The #FEMSmicroBlog welcomes external bloggers, writers and SciComm enthusiasts. Get in touch if you want to share your idea for a blog entry with us! |
